Structural models of biomolecules and complex biomolecular assemblies are required to answer fundamental questions related to physiology and pathophysiology. They also form the basis for developing drugs and optimizing protein properties in biotechnology.
However, the flexible and dynamic nature of biomolecules and complex biomolecular assemblies – which is essential for many cellular functions in living organisms – poses a major challenge for experimental methods used to determine high-resolution structural models as to time resolution and authenticity of the measurement conditions.
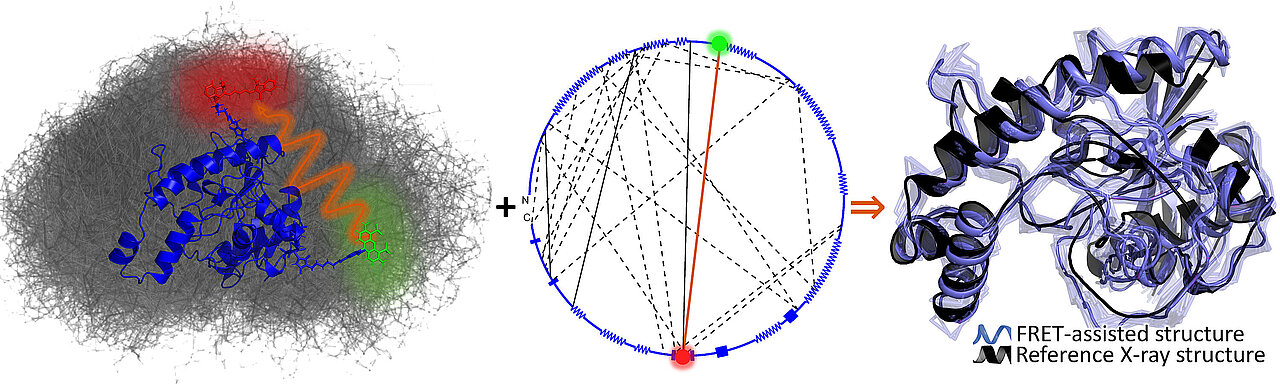
To master this challenge, scientists are carrying out fluorescence spectroscopy experiments. Using suitable FRET pairs (FRET spectroscopy) coupled to the protein as an optical reporter, these experiments provide information on distances between protein regions with a time resolution of approximately 10 nanoseconds (10-8 seconds) – similar to a strobe light. However, the information density obtained is not sufficient to create protein structural models with atomic resolution. Therefore, molecular simulations are used to generate many protein structural models at the atomic or coarse-grained level. Nevertheless, it is often unclear which of the models generated corresponds to reality.
To tackle this problem, Prof. Dr. Claus Seidel (Institute of Molecular Physical Chemistry) and Prof. Dr. Holger Gohlke (Institute of Pharmaceutical and Medicinal Chemistry) and their teams have now developed solutions. Their new quantitative integrative approach combines fluorescence spectroscopy with molecular simulations. After extensive preliminary work, they succeeded in creating a tool for the automated design of an optimally informative network of FRET pairs, developing new algorithmic approaches for combining molecular simulations and fluorescence experiments, and establishing, for the first time, an innovative method for the quantitative accuracy assessment of structural models created in this way.
These methods allow for planning FRET experiments efficiently and creating structural models of proteins with multiple conformational states with near-atomic resolution. Results for the enzyme T4 lysozyme can be accessed in the worldwide protein database (PDB-dev ID: PDBDEV_00000044). The approaches are available as open source software and on a web server. The FRET experiments are highly sensitive and can be used in natural conditions (in solution or in cells).
Contact
Publication
Dimura, M., Peulen, T.O., Sanabria, H., Rodnin, D., Hemmen, K., Seidel, C.A.M.*, Gohlke, H.*
Automated and optimally FRET-assisted structural modeling.
Nature Comm. 2020, 11, 5394.